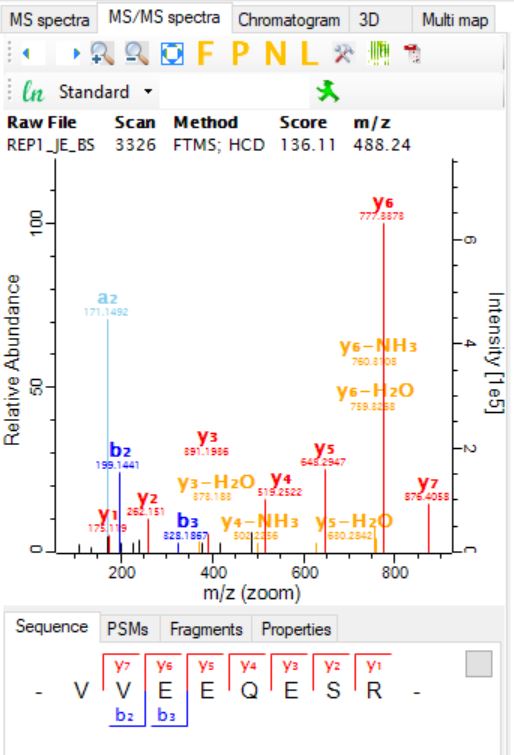
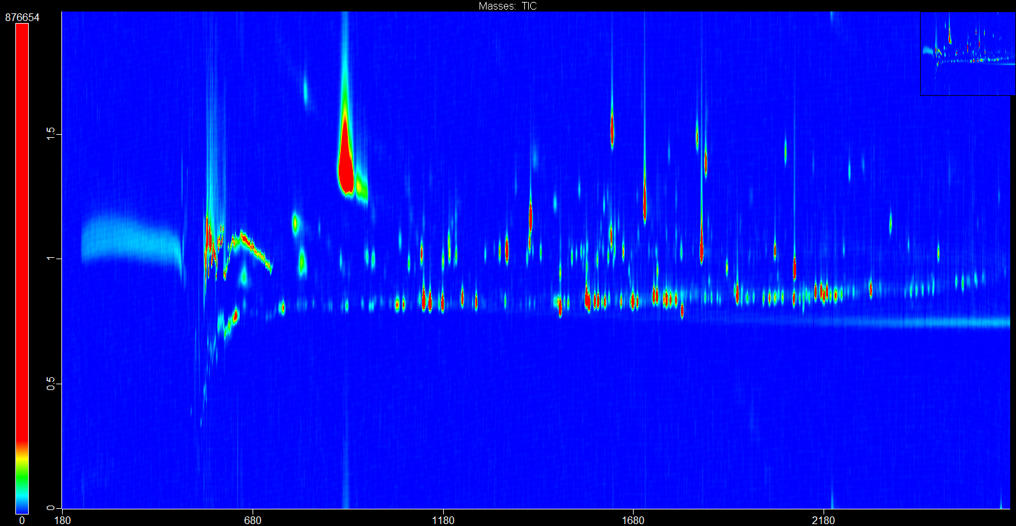
In collaboration with local clinics and biobanks, human-derived patient samples are characterized in an unbiased and global manner, in an effort to obtain information regarding the quantity of patient RNA, proteins, and metabolites. This information is then processed using bioinformatic methodologies to allow for the identification of disease-associated molecules (biomarkers) or novel disease states after comparison to healthy or control patients. Ultimately, multi-omics is a nascent, interdisciplinary field of bioengineering that acknowledges disease-associated biomarkers can simultaneously exist as DNA, RNA, proteins, and metabolites. By considering more than one class of endogenous molecules, bias is further reduced and previously unknown or unexpected interactions can be identified, allowing for deeper insight into the mechanisms driving disease.