Research Interests
- Develop novel biosensors and bioinstrumentations for biomedical research, drug development and disease diagnosis
- Label-free optical and electrochemical sensing for chemical and biological targets
Research Thrusts
Research Thrust 1: Functional Plasmonic Imaging of Molecular and Cellular Activities
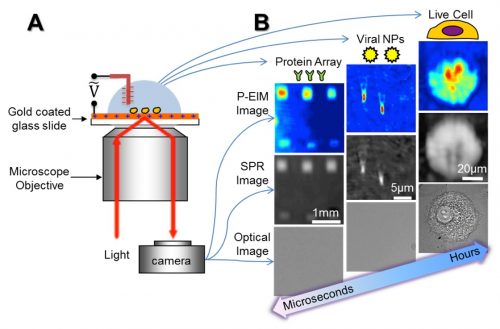
A major direction of my research is on plasmonic-based imaging technology, and the application toward chemical and biological targets. My lab developed multi-functional plasmonic-based electrochemical impedance microscopy (P-EIM, Science 2010). P-EIM uses objective or prism coupled distortion-free surface plasmon resonance microscopy (SPRM, PNAS 2010), to map electrochemical current and surface impedance of samples, in addition to regular bright field, dark field and fluorescence imaging. P-EIM can measure molecular absorptions, electrochemical reactions, electrical impedance, and cellular activities with high spatial and temporal resolution. Our lab demonstrated broad applications of P-EIM, including detection of small molecules, proteins, DNA, virus and bacteria, mapping single neuron cell signals, single nanoparticle catalytic events, 2D material properties and detection of trace explosives. Recently, we invented Plasmonic Scattering Microscopy (PSM, Nat. Meth. 2020) and Evanescence Scattering Microscopy (ESM, Nat. Commun. 2022) pushed the detection limit to single proteins. These results are transformative and lead to new tools for biomedical research, drug discovery, and diagnostics applications. This research thrust is supported by NIH, NSF, Keck and Moore foundation.
- Optical imaging of size, charge, mobility and binding of single proteins Protein analysis is essential to the understanding of molecular scale processes in living systems, the diagnosis of diseases based on molecular biomarkers, and the treatment of diseases with drugs. The basic tasks of protein analysis include detecting a protein, identifying it and determining its interactions with other proteins or molecular ligands. The most used technologies to perform these tasks are gel and capillary electrophoresis, Western Blot (WB) and enzyme linked immunosorbent assay (ELISA). These technologies identify proteins based on a protein’s charge, size, and specific binding to antibodies. For molecular interaction analysis, surface plasmon resonance and other detection technologies are the current choices. Although ubiquitous in both research labs and industry, these platforms must be combined to provide complete analysis of proteins, which is complicated and time consuming. In addition, they lack single molecule analysis capability required for studying heterogenous processes and for achieving precision diagnosis, especially for low volume samples. The project aims to develop one detection platform that can perform the key functions of above technologies with single molecule detection capability. We are using plasmonic imaging coupled with single protein oscillator to image single proteins without labels, measures the size, charge, and mobility of each protein simultaneously, identifies the protein based on its specific binding to antibodies, and quantifies its interactions with other proteins in real time. The project is currently funded by NIH (R01GM140193, 09/01/2022-08/31/2026).
- Quantitative label-free imaging of cellular activities in cells Electrical activities in cells play key roles in many important biological processes, including brain signal processing, cardiac functions, wound healing and other developmental processes. Currently, the most widely used experimental tool for studying cellular electrical activities measures a local electrical current or voltage with a microelectrode or micropipette. While sensitive, it lacks spatial resolution and is invasive. This project develops P-EIM for label-free imaging of electrical signals in cells. The microscope converts an electrical signal to a plasmonic signal, which is imaged optically. Building on the success of substantial preliminary studies (ACS nano 2019, Angew. Chem. 2017), we develop a new capability of P-EIM for studying cellular electrical activities and establish key applications. The success of this project will lead to a new tool for studying electrical activities in cells with temporal and spatial resolutions that are not possible with the existing technologies. This new label-free imaging tool will provide new insights into the roles of electrical activities in biological processes, and a new method for screening drugs targeting neuronal and cardiac functions, wound healing and other develop-mental processes. The project is currently funded by NIH (2R01GM107165, 09/01/2018-08/31/2023).
Research Thrust 2: Novel Methods to Quantify molecular interactions
Measuring molecular interactions of proteins is critical for understanding protein functions and cellular processes, for discovery and validating biomarkers, and for developing and screening drugs. In particular, membrane proteins play key roles in many cellular functions and are the largest class of drug targets. Most popular methods for measuring membrane protein interaction kinetics involve extraction and purification of membrane proteins and stabilizing the proteins in an artificial lipid environment, which is not only time consuming and labor intensive, but also may introduce bias due to the loss of the native cellular microenvironment. Measuring the kinetics of small molecule binding to protein receptors and biochemical reactions of proteins, such as post-translational protein modifications, is a basic task in the understanding of diseases, discovering of diagnosis biomarkers, and screening of drugs. Various label-free techniques have been developed, but their sensitivity decreases with the molecular mass, which makes it challenging to detect small molecules and biochemical reactions. To solve these hurdles, my lab invented a few novel methods to quantify small molecule and membrane protein binding kinetics based on different transduction mechanisms, including SPRM (Nat. Chem. 2012, & Sci. Adv. 2014) charge-based optical detection (Chem. Sci. 2014, ACS Sen. 2021), nano-oscillator (Nano Lett. 2014, Anal. Chem. 2019), measure binding induced cell membrane deformation (Sci. Adv. 2015), and near critical angle reflection imaging. We are actively working with a local instrumentation company, Biosensing Instrument (BI), to commercialize these technologies. This research thrust is supported by NIH, Amgen and Genentech.
Active projects:
- Critical angle reflection imaging (CARi) for label-free quantification of molecular interactions The project aims to develop critical angle reflection imaging (CARi) as a breakthrough technology for in-situ cell-based studies of membrane protein binding interaction kinetics to advance the field of biomarker discovery and drug development. CARi builds upon surface plasmon resonance imaging (SPRi), acquiring many of its unique advantages, but overcoming many of its limitations. CARi uses an optical configuration similar to SPRi that measures light reflected from below the sensing surface, which is sensitive to molecular bindings induced refractive index changes above the sensing surface. This enables CARi to detect molecular interaction label-free and in real-time. Compared to SPRi, CARi exhibits several distinct technological advances, including a greater sensitivity and vertical detection range for measuring entire cell surfaces, simultaneous fluorescence compatibility for orthogonal validation, broader wavelength of light selection, convenient use of glass-based surface chemistries, and simple low-cost glass sensor chips. My lab invented CARi (Nat. Commun. 2021) and is working with BI to develop a commercial prototype multi-functional CARi instrument. We also collaborate with potential customers in biomedical research (Profs. Joshua LaBaer) and pharmaceutical industry (Pfizer) to validate CARi performance and develop key applications. The success of this project will enable ultra-high sensitivity for label-free kinetic quantification of small molecule interactions on membrane proteins with single-cell resolution and permit simultaneous fluorescence imaging for orthogonal validation. This powerful capability of label-free in-situ cell-based kinetic binding analysis is greatly needed for expediting biomarker discovery, disease diagnosis and drug screening. The project is funded by NIH (R42GM143986, 08/15/2021 – 02/14/2024).
- A charge sensitive optical detection system (CSOD) for high-throughput study of small molecules The technology is not only sensitive to small molecules, but also compatible with the standard microplate technology, which is particularly suitable for high-throughput applications. The working principle was established and validated with the support of NIH IMAT (Innovative Molecular Analysis Technologies for cancer research) grants (R21 and R33). Currently, my lab works with BI to develop a commercially viable prototype system. We will continue collaborations with potential customers in biomedical research and pharmaceutical industries for testing samples relevant to their interest and validating the performance usability of the developing system. The success of the project will lead to a new high-throughput screening technology for measuring molecular interactions, particularly small molecule interactions with proteins, and post-translational modifications of proteins. These processes are highly important for biomarker discovery, disease diagnosis and drug screening. The project is funded by NIH (R44GM139535, 05/18/2021 – 03/31/2023)
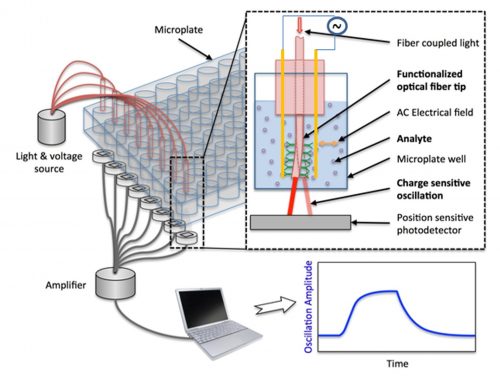
Charge sensitive optical detection (Chem. Sci. 2014, Anal. Chem. 2016, ACS Sen. 2020)
- A virion-display oscillator Array for quantification of transmembrane protein binding kinetics Transmembrane proteins, such as G-protein-coupled receptors (GPCRs), are critical for many cellular functions, and are also the popular drug targets for various diseases. For both understanding cellular functions and drug development, it is necessary to measure their binding activities with molecular ligands and drug candidates. In collaboration with Drs. Heng Zhu and Prashant Desai at Johns Hopkins University, this project addresses both challenges with a virion oscillator technology (#42 JACS 2018). Human GPCRs are displayed on the viral envelopes of human herpes simplex virus-1 (HSV-1), which removes the need of extraction, purification, and reconstitution of the transmembrane proteins. Each virion is then tethered to a sensor chip with a flexible polymer linker to form an oscillator. By applying an alternating electric field to the chip, the virion oscillates, and the oscillation amplitude is tracked in real-time with sub-nanometer precision using plasmonic imaging. Upon binding of ligands or drugs to the GPCRs on the virion envelopes, the oscillation amplitude changes, and from which binding kinetics and affinity are quantified. The goal of this project is to transform the technology into a powerful high-throughput platform for studying cellular functions of membrane proteins, and quantifying binding of large and small molecule drugs with any types of membrane proteins. The project is funded by NIH IMAT program (R33CA235294, 03/01/2020 02/28/2023)
Research Thrust 3: Optical Imaging Based Medical Diagnostic Methods and Devices
The third research thrust in my lab is developing novel diagnostic methods to solve unmet clinical needs. Most of these needs were requested or identified with help from clinical doctors, and we are working with them to develop practical solutions using the toolbox in our hand. We have three active projects in this area, funded by NIH (R01, pending R61) and a Mayo Clinic seed grant.
Active projects:
- Point-of-care antimicrobial susceptibility testing based on simultaneous tracking of multi-phenotypic features of single bacterial cells Antibiotic resistance has become a significant public health threat. To combat the problem, a rapid pathogen identification (ID) and antimicrobial susceptibility testing (AST) technology is needed to provide timely diagnosis of resistant infections and to deliver accurate antibiotic treatment at primary health-care settings, including hospitals and point-of-care (POC). The project aims to develop a point-of-care AST (POCAST) technology based on a large-image-volume microscopy technique that enables direct detection of individual bacterial cells in clinical samples without culturing or pathogen isolation, and a machine-learning model that allows fast determination of pathogen and susceptibility. This project is focus on 1) developing the large-image-volume microscopy and machine learning model for simultaneous tracking of multi-phenotypic features of single bacterial cells directly in patient urine sample, and performing rapid automatic pathogen ID and AST for UTIs; 2) building prototype instrument, and 3) validating the instrument for UTIs using large scale clinical samples. Successful development and validation of the technology will enable precise antibiotic prescription on the same day of patient visit. The project is carried out by a multidisciplinary team with expertise in biosensors (my lab), microbiology and infectious diseases (Dr. Haydel at Biodesign BB Center), biomedical instrument development and production (BI), and clinical testing (Dr. Grys at Clinical Microbiology Laboratory, Mayo Clinic). The project is funded by NIH (1R01AI138993, 07/25/2018 – 06/30/2023).

Rapid Antimicrobial Susceptibility Test (ACS Nano 2016, Anal. Chem. 2017, 2018, 2019, small 2020)
- Digital immunoassay for rapid and precise detection of blood biomarkers Cardiac troponin has become an essential biomarker in the diagnosis and risk stratification of patients presenting with acute coronary syndrome (ACS). In collaboration with Dr. Eric Yang (department of cardiovascular medicine) and Dr. Christine Snozek (director of clinical chemistry) at Mayo Clinic Arizona, we are developing a point-of-care (POC) hs-cTn assay that requires minimal blood sample, and is rapid, accurate, sensitive, and portable. We have developed a novel imaging-based digital immunoassay technology that can rapidly detect protein biomarkers using real time counting of antibodies labeled with gold nanoparticles (ACS Nano 2019, ACS sensor 2020). An initial proof of concept model for rapid hs-cTn detection using this technology has been published (ACS sensor 2021) using samples obtained from ACS patients at Mayo Clinic Arizona (MCA). We are developing a POC prototype device that will be clinically tested in ACS patients, to obtain sufficient data for an NIH R01 grant proposal. The project is currently funded by Mayo Clinic Arizona Cardiovascular Research Grant Award, FP00115547, 08/01/2021 – 01/31/2023.
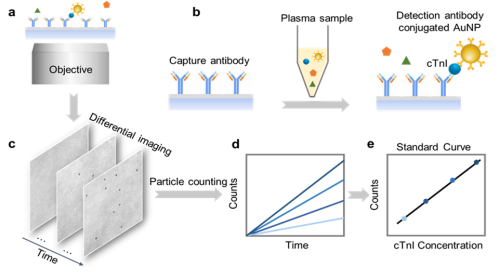
Imaging-based Digital Immunoassay (ACS Nano 2019, ACS sensor 2020, 2021)
Active Sponsored Research Projects
- Shaopeng Wang,* Optical imaging of size, charge, mobility and binding of single proteins, NIH NIGMS R01GM140193, 09/01/2022- 08/31/2026
- Shaopeng Wang,* Critical angle reflection imaging for label-free quantification of molecular interactions, Biosensing Instrument Inc., (VIA NIH NIGMS R42GM143986) 08/15/2021 – 02/14/2022
- Shaopeng Wang,* Development and Validation of a Rapid, Point-of-Care Digital Immunoassay for High Sensitivity Cardiac Troponin Detection, Subaward of Mayo Clinic Arizona Cardiovascular Research Grant Award, 08/01/2021 – 01/31/2023
- Thomas, Mary Laura (CoI)Wang, Shaopeng (CoI)Wu, Teresa (CoI)Forzani, Erica (PI), Partnership for Innovation – Avoiding Kidney Injuries with Evidence – Based Smart Technology, NSF 2122901, 09/01/2021 – 08/31/2024
- Shaopeng Wang,* Development of a charge-sensitive optical detection system for high-throughput study of small molecule binding kinetics, Biosensing Instrument Inc., (VIA NIH NIGMS R44GM139535), 5/18/2021 – 3/31/2023
- Shaopeng Wang,* (Nongjian Tao was original PI) A Virion-Display Oscillator Array and Detection Platform for Quantification of Transmembrane Protein Binding Kinetics, NIH NCI IMAT R33CA235294 03/01/2020 – 02/28/2023
- Shaopeng Wang,* Quantitative label-free imaging of electrical activities in cells, NIH NIGMS 2R01GM107165, 09/01/2018-08/31/2022
- Shaopeng Wang,* Shelley Haydel, (Nongjian Tao was original PI) Point-of-care antimicrobial susceptibility testing based on simultaneous tracking of multi-phenotypic features of single bacterial cells, NIH NIAID 1R01AI138993-01 07/25/2018 – 06/30/2023
Completed Sponsored Research Projects
- Shaopeng Wang,* (Nongjian Tao was original PI) Measuring small molecule interactions with membrane proteins on single cells via detecting nanometer scale membrane deformations, NIH NIGMS R01GM124335, 07/01/2017 – 04/30/2022
- Shaopeng Wang,* Direct quantification of binding kinetics between phage and mRNA displayed peptide, Genentech Inc. AGR 11/05/18, 10/30/2018 – 10/29/2021
- Shaopeng Wang,* (Nongjian Tao was original PI) Nano-Oscillator Arrays for Sensitive Plasmonic Detection of Molecular Interaction, Biosensing Instrument Inc., BIG126720 (VIA NIH NIGMS R44GM126720), 5/1/2018 – 10/31/2021
- Shaopeng Wang,* Nongjian Tao, An Integrated Microarray Printing and Detection System, Biosensing Instrument Inc., (Via NIH NIGMS 1R44GM114951-01), 4/1/2015-3/31/2021
- Nongjian Tao,* Shaopeng Wang, Charge sensitive optical detection for high-throughput study of small molecules. NIH NCI IMAT R33CA202834 08/01/2016 – 07/31/2020
- Shaopeng Wang,* Nongjian Tao, Quantitative label-free imaging of membrane protein interaction kinetics on cells, NIH NIGMS 1R01GM107165, 07/01/2014-06/30/2018
- Nongjian Tao,* Shaopeng Wang, Label-free imaging and tracking of single protein-protein interaction, Moore Foundation, 04/12/2013-07/31/2016
- Nongjian Tao*, Shaopeng Wang, Electrochemically-Enhanced Plasmonic Imaging for Quantitative Proteomics, Biosensing Instrument Inc.(Via NIH NIGMS 1R44GM106579), 04/01/2013-03/31/2017
- Nongjian Tao,* Shaopeng Wang, Charge sensitive optical detection for high-throughput study of small molecules, NIH NCI IMAT R21CA173205, 09/01/2012 – 08/31/2015
- Nongjian Tao,* Shaopeng Wang, IDBR: Plasmonic-based electrochemical impedance microscopy for studying molecular binding and cellular processes NSF #1151005, 02/01/2012 – 01/31/2016
- Nongjian Tao,* Shaopeng Wang, A Self-Assembled Nanomechanical System (NMES) for Molecular Detections, W. M. Keck Foundation, 07/01/2011-06/30/2016
- Shaopeng Wang,* Evaluate P-EIM technology for membrane protein binding affinities measurement, Amgen 2012577119 11/13/12 – 05/12/15
- Nongjian Tao,* Shaopeng Wang, Plasmonic Mapping of Ion Channel Activities in Single Cell, NIH R21 1R21DA033839-01, 04/01/2012 – 03/31/2015
- Nongjian Tao,* Shaopeng Wang, Measure Protein Interaction at Scale, Piper Bridge Award (Virginia Piper Foundation), 04/01/2012-06/30/2013
- Shaopeng Wang,* Nongjian Tao, A Multi-functional Optical Impedance Microscope for Live Cell Imaging NIH R21 RR026235 (8R21GM103396), 05/15/2010-02/28/2013
* Principal Investigator